It was great to talk about reproducible workflows for @riotscienceclub @riotscience_wlv. You can watch the recording below, but if you don't want to listen to me talk for 40 minutes, I thought I would summarise my talk in a thread:
Thank you again @JamesEBartlett for a fantastic talk (with a really nice personal touch) on reproducible workflows!
— RIOT Science Club Wolverhampton (@riotscience_wlv) February 16, 2021
Thanks especially for the co-leads @IMLahart for co-hosting and @DrManiBhogal for nabbing James!
Slides: https://t.co/CNqxzOhch1
Video: https://t.co/YjHEHuRJlz

Why are lunch breaks important for #code?
— Dr Rebecca Hirst (@HirstRj) February 11, 2021
If you can't remember what your variable names refer to after lunch, you sure as hell won't remember in 3 months.


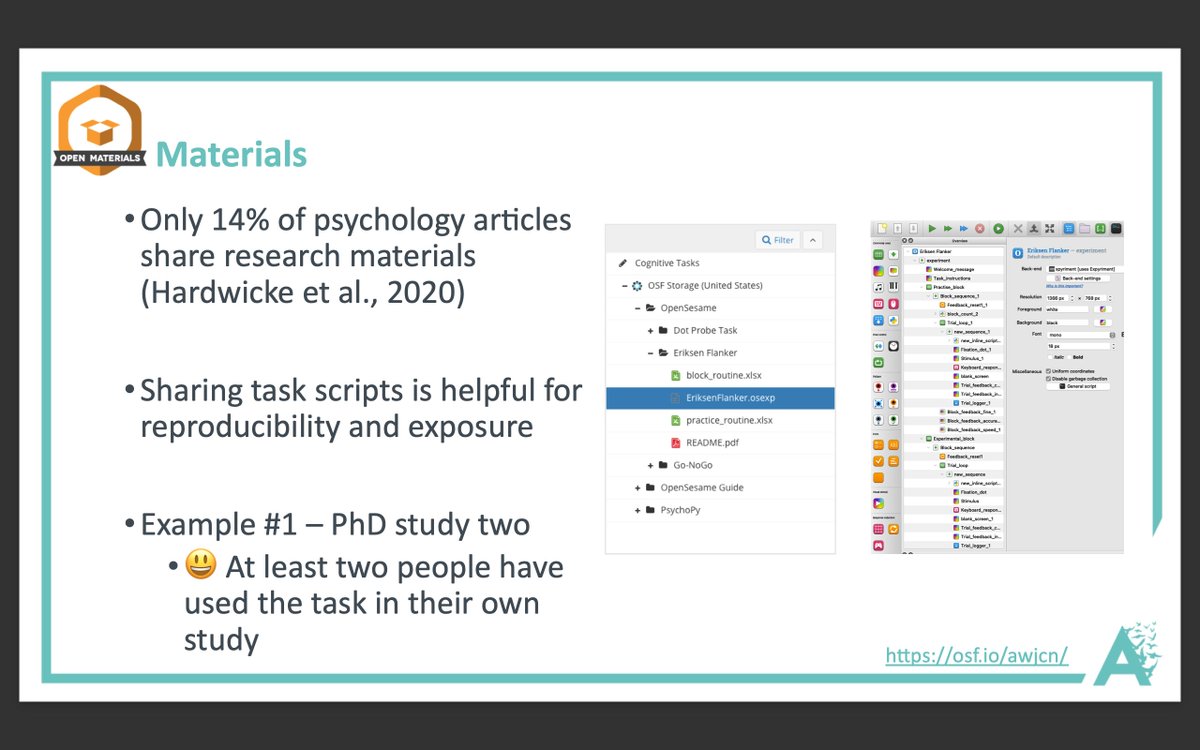
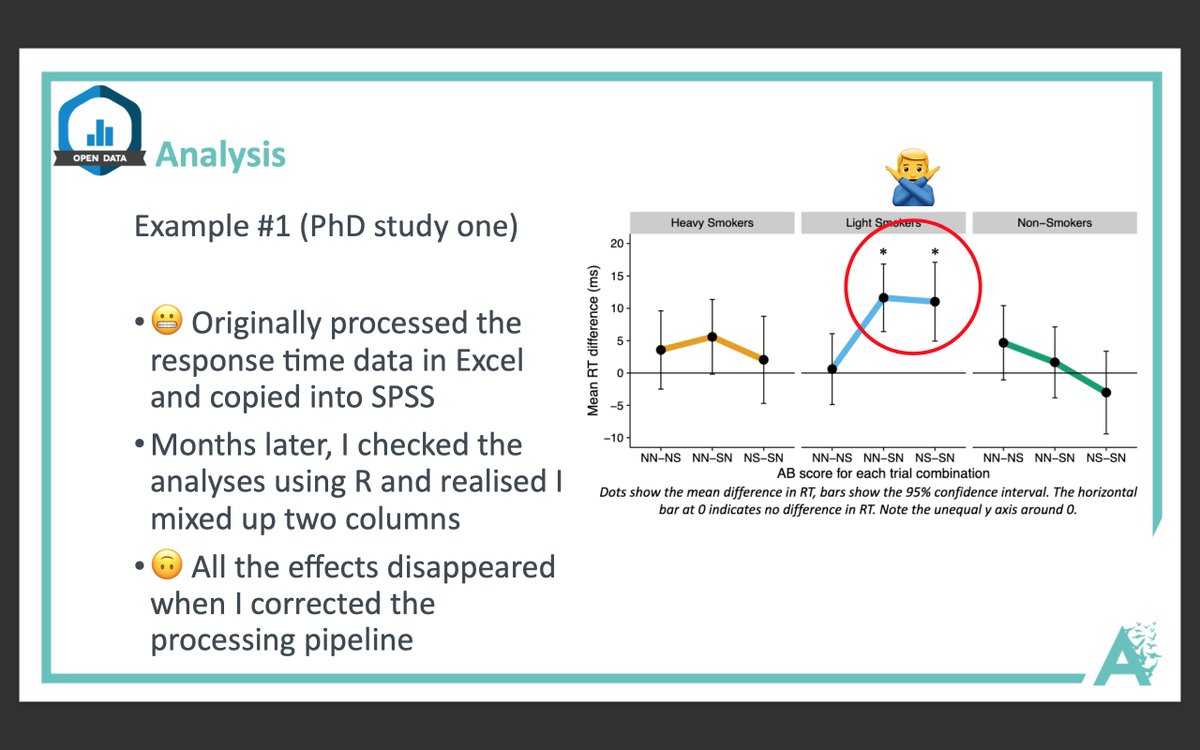
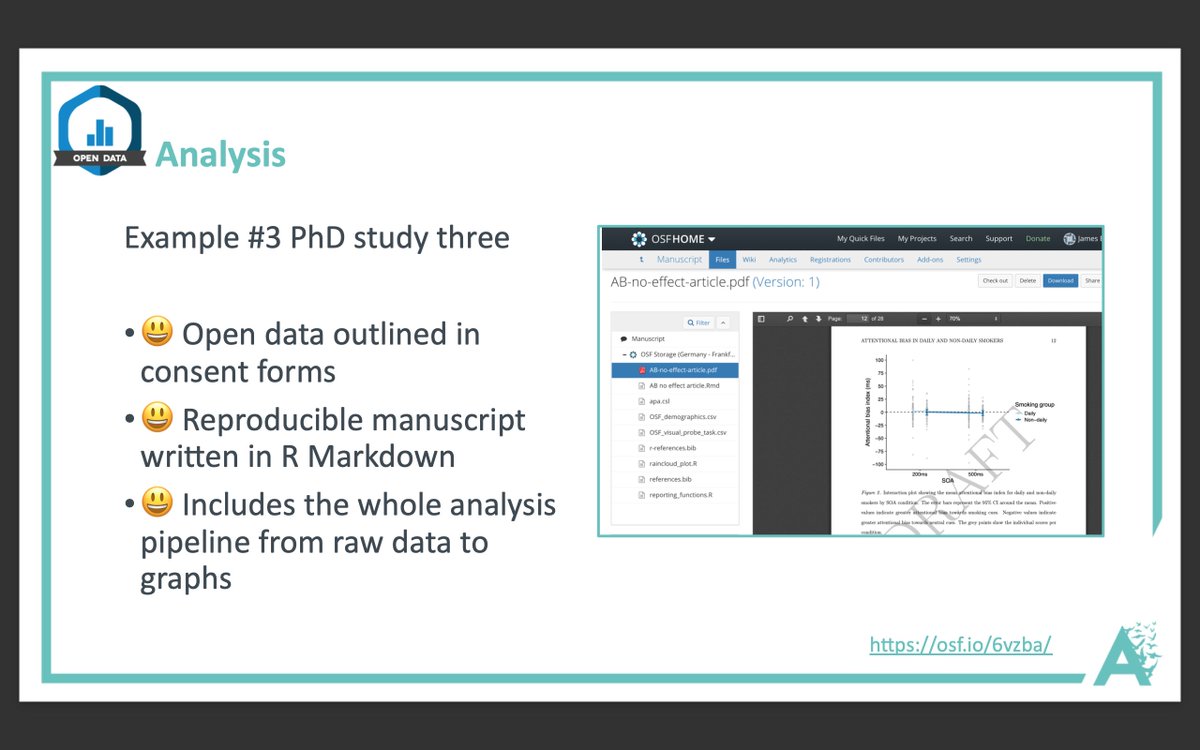

More from Science
Variants always emerge, & are not good or bad, but expected. The challenge is figuring out which variants are bad, and that can't be done with sequence alone.
Feels like the next thing we're going to need is a ranking system for how concerning "variants of concern\u201d actually are.
— Kai Kupferschmidt (@kakape) January 15, 2021
A lot of constellations of mutations are concerning, but people are lumping together variants with vastly different levels of evidence that we need to worry.
You can't just look at a sequence and say, "Aha! A mutation in spike. This must be more transmissible or can evade antibody neutralization." Sure, we can use computational models to try and predict the functional consequence of a given mutation, but models are often wrong.
The virus acquires mutations randomly every time it replicates. Many mutations don't change the virus at all. Others may change it in a way that have no consequences for human transmission or disease. But you can't tell just looking at sequence alone.
In order to determine the functional impact of a mutation, you need to actually do experiments. You can look at some effects in cell culture, but to address questions relating to transmission or disease, you have to use animal models.
The reason people were concerned initially about B.1.1.7 is because of epidemiological evidence showing that it rapidly became dominant in one area. More rapidly that could be explained unless it had some kind of advantage that allowed it to outcompete other circulating variants.
I'm going to answer the question so many people have been asking this week:
WHAT IS PROJECT X???
Here's the definitive thread to tell you - and show you -precisely what Project X is
Grab a drink, sit down with me and let's #TalkLiberation
<3
1/?
"Project X" is actually called "PanQuake".
Pan means "all". Quake is the huge effect our voices can have when our communications are uncensored and when we have access to brand new functionality that *enhances* our social reach, rather than diminishes it
Here's our logo:
2/?

You can follow the fledgling official PanQuake Twitter account here: @pan_quake and see our super cool new website here: https://t.co/F7wLSeM6aK
You can find our donation page here: https://t.co/VICFnsR0RX
Keep reading this thread to find out why we created it & what it is
3/?
SPOILER ALERT: Much of the content below this point is from my personal slides & speech notes from today's launch event. That stream got totally ruined by (big) tech problems, but I'm happy to report everything is turning out wonderfully
Not one single team member or guest left. We are all still here, smiling not crying, as we record this event and will get it out to you all very soon :)
— Suzie Dawson (@Suzi3D) January 17, 2021
I'm so proud of everyone, what an amazing crewhttps://t.co/RmE0BicIXF
Here are some of our most high profile & dedicated public advocates for PanQuake - many of whom were scheduled to appear at our launch. All of whom stuck around for hours, to do a prerecord of the event, which is being edited, processed & uploaded for you as I write this.
5/?