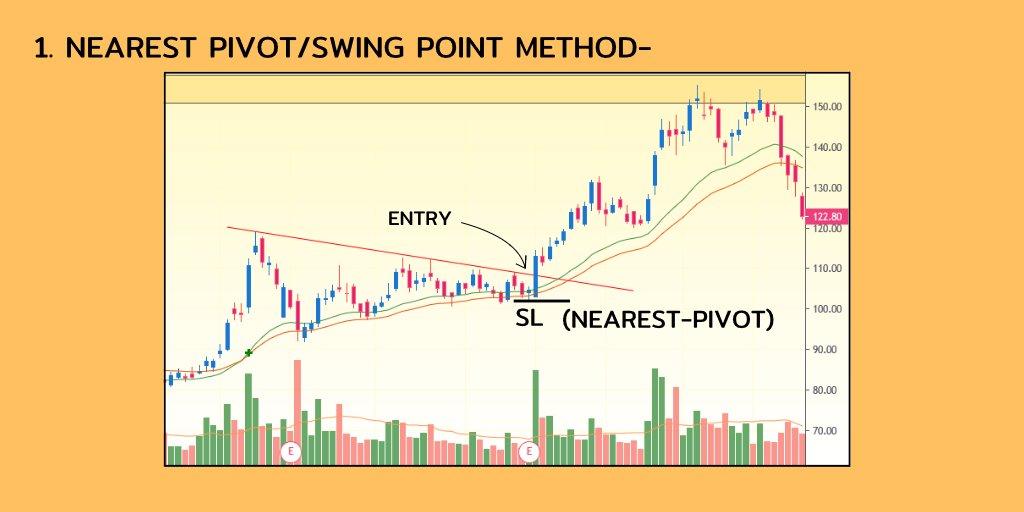
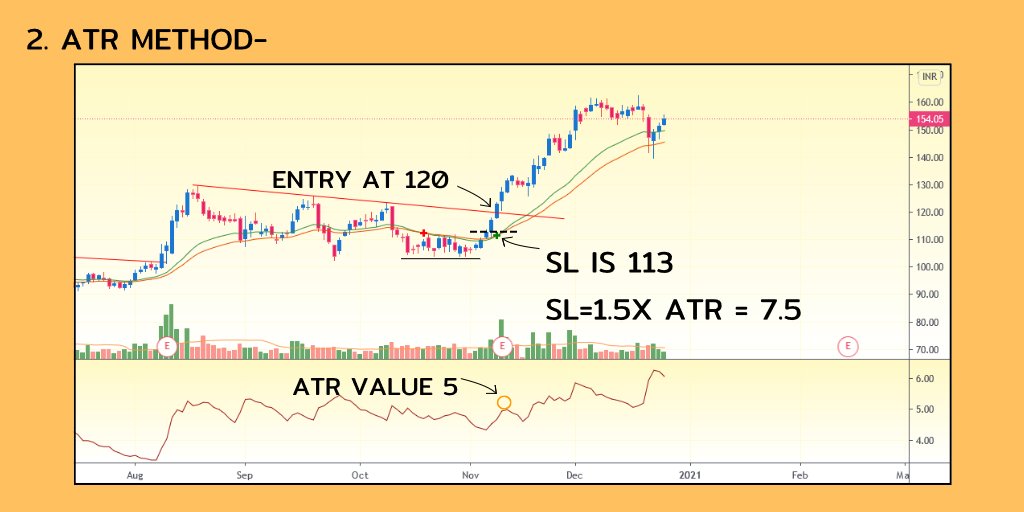
People often ask why structural variants (SV) are so important and why $BNGO is laser focused on revolutionizing the way they are detected in the clinic. Doesn’t the success of $PACB and $ILMN show that sequencing and the small variants it detects are what matters?
More from Trading
1/ Feels like a good time to tell the story of how I went from broke to a millionaire to broke again in 2017/18 again...
Yesterday was brutal for some people...
Losing life-changing money sucks, losing any money sucks...you can chase the market or you can change your strategy.
2/ The original thread is gone but you can read it here.
https://t.co/cLLNs75rB0
tl;dr
- Traded $32k to $1.2m
- Thought I was a genius
- Made poor investments
- Didn't conserve capital
- Peaked at 150 BTC
- Lost nearly all of it
2 weeks from losing my house + no income. Oops.
3/ I am going to assume you are in it for the money rather than the tech. Yeah, you might Tweet about the amazing blockchaining of cross-border payments and oracles yadda yadda...really, you are in it to make money.
If you are really in it for the tech, go and build something.
4/ Okay, so if you want to make money, trading is super hard, you are trading against:
- Better traders than you
- People who can move markets
- Unknown information
And if you are trading with leverage you might blow up your account with the volatility.
5/ If you are not trading, you are investing. Okay, so what are you investing in?
I made the decision that the crypto with the best opportunity of existing in 10 years is #Bitcoin:
- Solves a genuine problem
- The right tech
- A proven track record
Yesterday was brutal for some people...
Losing life-changing money sucks, losing any money sucks...you can chase the market or you can change your strategy.
2/ The original thread is gone but you can read it here.
https://t.co/cLLNs75rB0
tl;dr
- Traded $32k to $1.2m
- Thought I was a genius
- Made poor investments
- Didn't conserve capital
- Peaked at 150 BTC
- Lost nearly all of it
2 weeks from losing my house + no income. Oops.
3/ I am going to assume you are in it for the money rather than the tech. Yeah, you might Tweet about the amazing blockchaining of cross-border payments and oracles yadda yadda...really, you are in it to make money.
If you are really in it for the tech, go and build something.
4/ Okay, so if you want to make money, trading is super hard, you are trading against:
- Better traders than you
- People who can move markets
- Unknown information
And if you are trading with leverage you might blow up your account with the volatility.
5/ If you are not trading, you are investing. Okay, so what are you investing in?
I made the decision that the crypto with the best opportunity of existing in 10 years is #Bitcoin:
- Solves a genuine problem
- The right tech
- A proven track record
You May Also Like
@EricTopol @NBA @StephenKissler @yhgrad B.1.1.7 reveals clearly that SARS-CoV-2 is reverting to its original pre-outbreak condition, i.e. adapted to transgenic hACE2 mice (either Baric's BALB/c ones or others used at WIV labs during chimeric bat coronavirus experiments aimed at developing a pan betacoronavirus vaccine)
@NBA @StephenKissler @yhgrad 1. From Day 1, SARS-COV-2 was very well adapted to humans .....and transgenic hACE2 Mice
@NBA @StephenKissler @yhgrad 2. High Probability of serial passaging in Transgenic Mice expressing hACE2 in genesis of SARS-COV-2
@NBA @StephenKissler @yhgrad B.1.1.7 has an unusually large number of genetic changes, ... found to date in mouse-adapted SARS-CoV2 and is also seen in ferret infections.
https://t.co/9Z4oJmkcKj

@NBA @StephenKissler @yhgrad We adapted a clinical isolate of SARS-CoV-2 by serial passaging in the ... Thus, this mouse-adapted strain and associated challenge model should be ... (B) SARS-CoV-2 genomic RNA loads in mouse lung homogenates at P0 to P6.
https://t.co/I90OOCJg7o

@NBA @StephenKissler @yhgrad 1. From Day 1, SARS-COV-2 was very well adapted to humans .....and transgenic hACE2 Mice
1. From Day 1, SARS-COV-2 was very well adapted to humans .....and transgenic hACE2 Mice
— Billy Bostickson \U0001f3f4\U0001f441&\U0001f441 \U0001f193 (@BillyBostickson) January 30, 2021
"we generated a mouse model expressing hACE2 by using CRISPR/Cas9 knockin technology. In comparison with wild-type C57BL/6 mice, both young & aged hACE2 mice sustained high viral loads... pic.twitter.com/j94XtSkscj
@NBA @StephenKissler @yhgrad 2. High Probability of serial passaging in Transgenic Mice expressing hACE2 in genesis of SARS-COV-2
1. High Probability of serial passaging in Transgenic Mice expressing hACE2 in genesis of SARS-COV-2!
— Billy Bostickson \U0001f3f4\U0001f441&\U0001f441 \U0001f193 (@BillyBostickson) January 2, 2021
2 papers:
Human\u2013viral molecular mimicryhttps://t.co/irfH0Zgrve
Molecular Mimicryhttps://t.co/yLQoUtfS6s https://t.co/lsCv2iMEQz
@NBA @StephenKissler @yhgrad B.1.1.7 has an unusually large number of genetic changes, ... found to date in mouse-adapted SARS-CoV2 and is also seen in ferret infections.
https://t.co/9Z4oJmkcKj

@NBA @StephenKissler @yhgrad We adapted a clinical isolate of SARS-CoV-2 by serial passaging in the ... Thus, this mouse-adapted strain and associated challenge model should be ... (B) SARS-CoV-2 genomic RNA loads in mouse lung homogenates at P0 to P6.
https://t.co/I90OOCJg7o