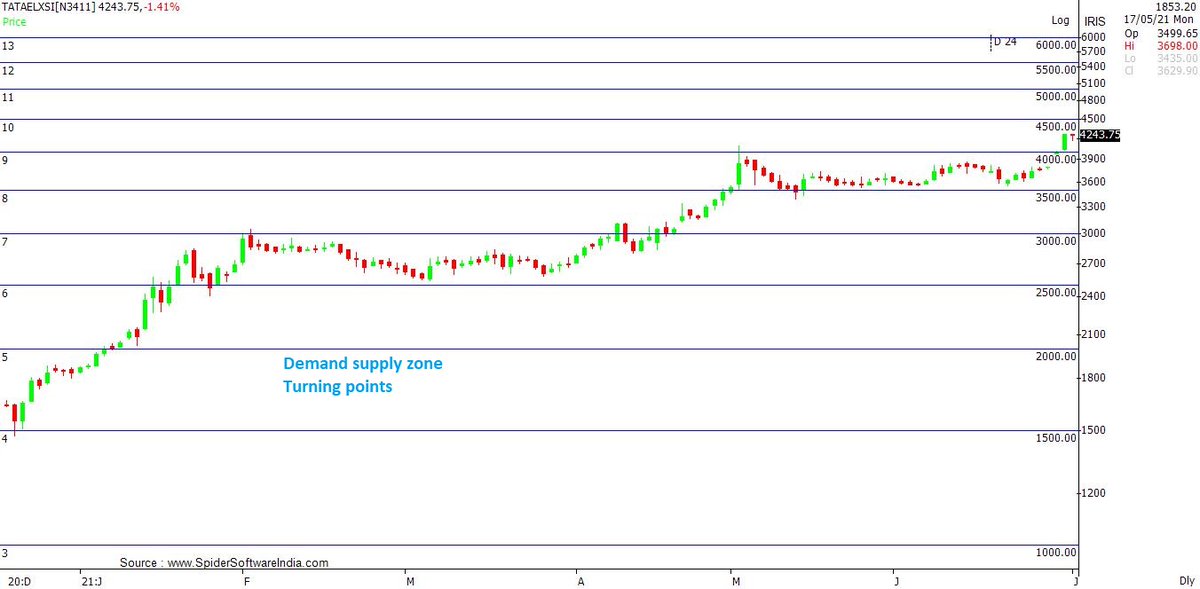
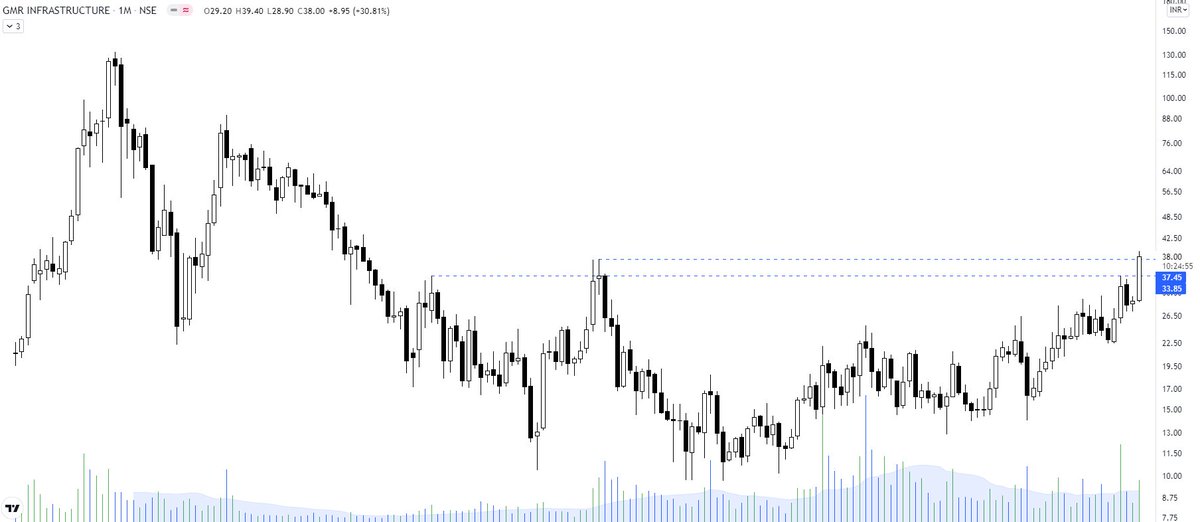
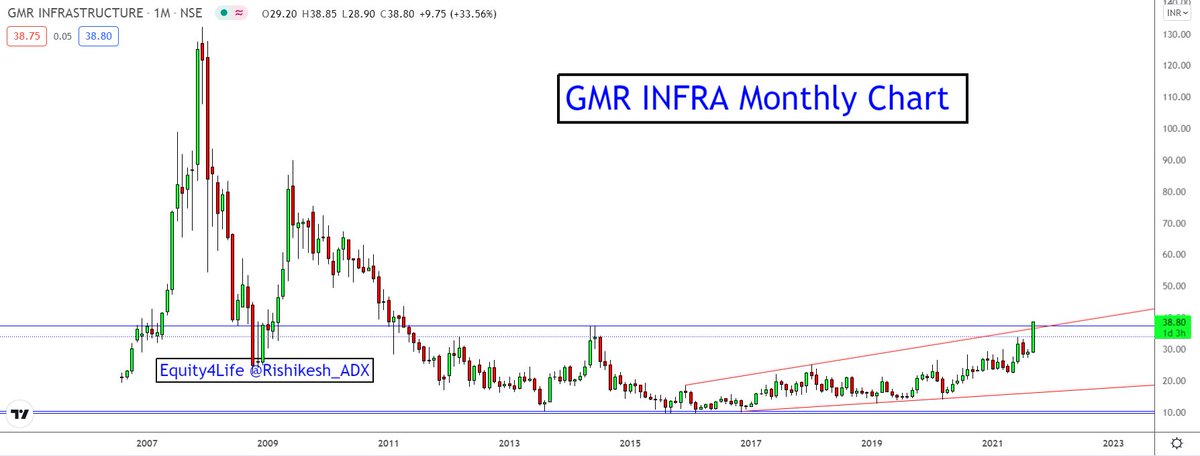
my next #BUY
#gmrinfra sl 22 on closing basis tgt 50
#Bullish #breakouts #outperformance #relativestrength
#investing #AhmedabadNest #Nishshkumar #India

More from NISHSHKUMAR JAANI
band walk start quick towards 800! https://t.co/DX5iHPePIC


#GUJGAS
— NISHSHKUMAR JAANI (@NishshkumarJaan) July 20, 2021
batting with 5% Risk for 30% Gains
entry abv 678 sl 647 tgt 894-Gone side ways for month now #Breakouts #relativestrength #outperformance #AhmedabadNest #investing #India pic.twitter.com/QWGSVJqOdK
More from Gmrinfra
#GMRINFRA
https://t.co/nY0plirqMe
https://t.co/nY0plirqMe
#GMRINFRA
— @TradeWise\u2122 \u26a1 (@charan_more) September 28, 2020
For the last 8-9 years, we have witnessed sideways trading range GMRINRA between 10-35 with multiple rounding bottom. are we missing the breakout timing of GMRINRA after one decade narrowed trading? pic.twitter.com/j8uxP3g5oT
You May Also Like
Keep dwelling on this:
Further Examination of the Motif near PRRA Reveals Close Structural Similarity to the SEB Superantigen as well as Sequence Similarities to Neurotoxins and a Viral SAg.
The insertion PRRA together with 7 sequentially preceding residues & succeeding R685 (conserved in β-CoVs) form a motif, Y674QTQTNSPRRAR685, homologous to those of neurotoxins from Ophiophagus (cobra) and Bungarus genera, as well as neurotoxin-like regions from three RABV strains
(20) (Fig. 2D). We further noticed that the same segment bears close similarity to the HIV-1 glycoprotein gp120 SAg motif F164 to V174.
https://t.co/EwwJOSa8RK
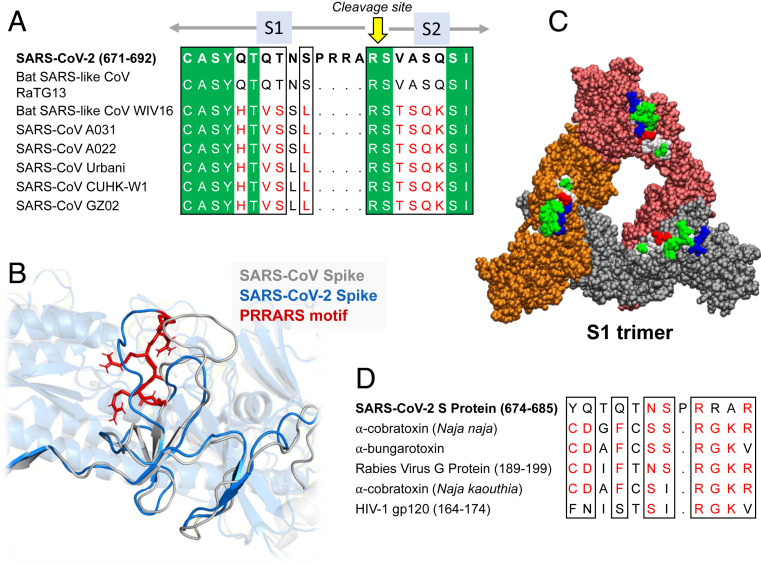
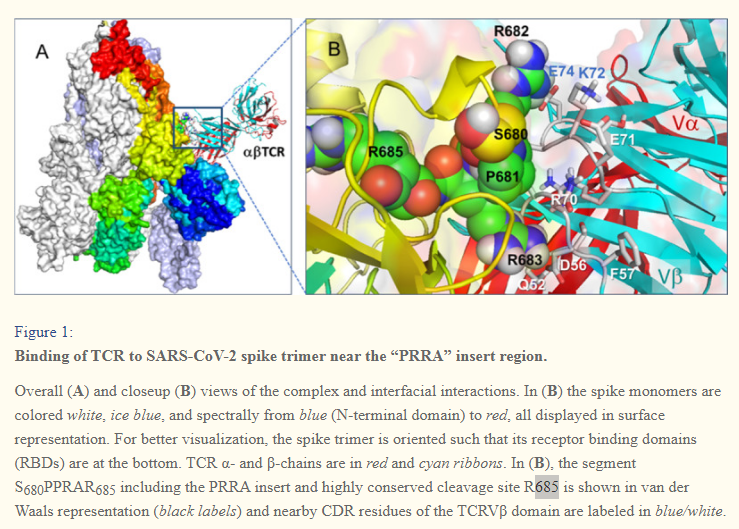
In (B), the segment S680PPRAR685 including the PRRA insert and highly conserved cleavage site *R685* is shown in van der Waals representation (black labels) and nearby CDR residues of the TCRVβ domain are labeled in blue/white
https://t.co/BsY8BAIzDa
Sequence Identity %
https://t.co/BsY8BAIzDa
Y674 - QTQTNSPRRA - R685
Similar to neurotoxins from Ophiophagus (cobra) & Bungarus genera & neurotoxin-like regions from three RABV strains
T678 - NSPRRA- R685
Superantigenic core, consistently aligned against bacterial or viral SAgs
Further Examination of the Motif near PRRA Reveals Close Structural Similarity to the SEB Superantigen as well as Sequence Similarities to Neurotoxins and a Viral SAg.
The insertion PRRA together with 7 sequentially preceding residues & succeeding R685 (conserved in β-CoVs) form a motif, Y674QTQTNSPRRAR685, homologous to those of neurotoxins from Ophiophagus (cobra) and Bungarus genera, as well as neurotoxin-like regions from three RABV strains
(20) (Fig. 2D). We further noticed that the same segment bears close similarity to the HIV-1 glycoprotein gp120 SAg motif F164 to V174.
https://t.co/EwwJOSa8RK

In (B), the segment S680PPRAR685 including the PRRA insert and highly conserved cleavage site *R685* is shown in van der Waals representation (black labels) and nearby CDR residues of the TCRVβ domain are labeled in blue/white
https://t.co/BsY8BAIzDa

Sequence Identity %
https://t.co/BsY8BAIzDa
Y674 - QTQTNSPRRA - R685
Similar to neurotoxins from Ophiophagus (cobra) & Bungarus genera & neurotoxin-like regions from three RABV strains
T678 - NSPRRA- R685
Superantigenic core, consistently aligned against bacterial or viral SAgs